YCN-RW: Bipartite Projections
YCN Random Walk
Implemented by me
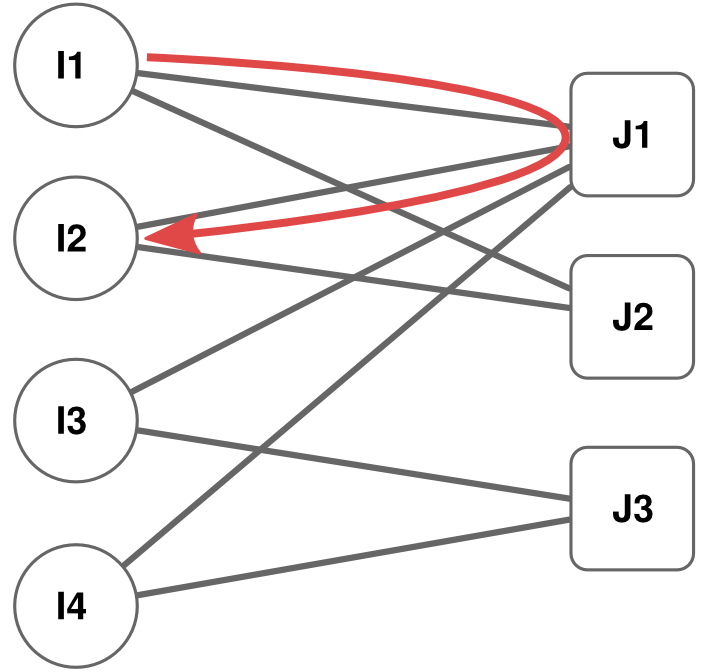
NEW VERSION! For the old implementation for the result reproducibility of the original paper, scroll below.
A custom Python library to perform bipartite network projection: transforming a bipartite network in a weighted unipartite network. Methods implemented:
- Simple projections (count, cosine, pearson, euclidean, jaccard);
- Hyperbolic projection [1];
- YCN Random Walks [2];
- ProbS [3], HeatS, & Hybrid [4].
Simply place the python script in your path or work folder. Each method requires you to pass a networkx graph and the list of nodes you want in your projection. It will return another networkx graph containing the projection and its weighted edges. An example of usage:
import networkx as nx
import network_map2 as nm2
G = nx.read_edgelist("/path/to/your/network")
nodes = nx.algorithms.bipartite.basic.sets(G)
rows = sorted(list(nodes[0]))
cols = sorted(list(nodes[1]))
Gp = nm2.simple(G, rows)
Note that YCN, ProbS, HeatS, and Hybrid can return asymmetric projections (where the i,j weight is different than the j,i weight). By default they do not, so you have to manually set the “directed” paramter to True:
Gp = nm2.ycn(G, rows, directed = True)
Moreover, Hybrid takes an additional mandatory parameter, l, which regulates how much you want to weight HeatS over ProbS. Setting it to 0.5 means you take the middle point between the two:
Gp = nm2.hybrid(G, rows, .5, directed = True)
Note that this is all implemented in numpy, scipy, sklearn and networkx, so you have to have those libraries to use these methods. Also, while most methods use sparse matrix representation, some cannot, thus for large networks you might not be able to store the result in memory.
For any inquiry, contact me at: michele.coscia@gmail.com
[1] Mark EJ Newman. Scientific collaboration networks. ii. shortest paths, weighted networks, and centrality. Physical review E, 64(1):016132, 2001c
[2] Muhammed A Yildirim and Michele Coscia. Using random walks to generate associations between objects. PloS one, 9 (8):e104813, 2014
[3] Tao Zhou, Jie Ren, Matus Medo, and Yi-Cheng Zhang. Bipartite network projection and personal recommendation. Physical Review E, 76(4):046115, 2007
[4] Tao Zhou, Zoltan Kuscsik, Jian-Guo Liu, Matus Medo, Joseph Rushton Wakeling, and Yi-Cheng Zhang. Solving the apparent diversity-accuracy dilemma of recommender systems.
Proceedings of the National Academy of Sciences, 107(10):4511–4515, 2010
LEGACY CODE! Only for reproducing the results in the paper.
This archive contains the code for utilizing the YCN-RW bipartite network projecting technique and for repeating the experiments of the paper “Using random walks to generate associations between objects” by Muhammed Yildirim and yours truly.
For any inquiry, contact me at: michele.coscia@gmail.com
The archive contains the following files:
– network_map.py: the Python library where the implementation is stored, along with other projection techniques that can be used (Cosine, Euclidean, Pearson, Jaccard and undirected Zhou, from the paper “Bipartite network projection and personal recommendation“).
– experiment.py: a sample Python script file that illustrate how to use the library and it is set up for verifying the experiments of the paper.
– experiment_movielens.py: another Python script file that illustrate how to use the library and it is set up for verifying the experiments of the paper.
– pyroc.py: utility Python script invoked by experiment.py to draw ROC curves and calculate the AUC. Created by Marcel Caraciolo on 2009-11-16.
– aid_bipartite: the bipartite network for the Aid dataset. It contains four tab-separated columns: entity1, entity2, type_of_entity1, type_of_entity2.
– aid_evalnet: the unipartite observed network for the Aid dataset. It contains three tab-separated columns: entity1, entity2, weight_of_unipartite_link.
– congress_bipartite: the bipartite network for the Congress dataset. It contains four tab-separated columns: entity1, entity2, type_of_entity1, type_of_entity2.
– congress_evalnet: the unipartite observed network for the Congress dataset. It contains three tab-separated columns: entity1, entity2, weight_of_unipartite_link.
– ipums_bipartite: the bipartite network for the IPUMS dataset. It contains four tab-separated columns: entity1, entity2, type_of_entity1, type_of_entity2.
– ipums_evalnet: the unipartite observed network for the IPUMS dataset. It contains three tab-separated columns: entity1, entity2, weight_of_unipartite_link.
– onet_bipartite: the bipartite network for the O-NET dataset. It contains four tab-separated columns: entity1, entity2, type_of_entity1, type_of_entity2.
– onet_evalnet: the unipartite observed network for the O-NET dataset. It contains three tab-separated columns: entity1, entity2, weight_of_unipartite_link.
– movielens_bipartite: the bipartite network for the Movielens dataset. It contains four tab-separated columns: entity1, entity2, type_of_entity1, type_of_entity2.
– README: a file containing this description.
Requirements:
network_map.py requires that you installed the Numpy, Scipy and Scikit-learn Python libraries.
experiment.py additionally requires the Matplotlib Python library (it is required only for plotting the ROC curve and AUC calculation, so if you don’t want that, you can remove this requirement by commenting the relative lines, #23-33).
Usage instructions:
To perform YCN-RW on a bipartite network, or use any other implemented method in network_map.py, first put the network_map.py file in the directory where you will run your Python script. Then, import it as a module: “import network_map” at the beginning of the file. Then, you can call the method by calling “edges = network_map.ycn_edges_2(bipartite_edges, key_map, directed = True/False)“. The two parameters are:
– “bipartite_edges“: it has to be a dictionary. The key of the dictionary is the entity id. The value of the dictionary is the entity’s adjacency list, i.e. the list of ids of the entities this entity is connected to. Both entity types of the bipartite network has to be present as keys of the dictionary (see experiment.py lines 60-61).
– “key_map“: it has to be a dictionary. The key of the dictionary is the entity id. The value of the dictionary is 0 if the entity is of the type that has to be in the unipartite network, 1 otherwise (see experiment.py lines 58-59).
– “directed“: optional parameter. If True, then the x-y similarity will be different from the y-x similarity. Default: False.
The output is a dictionary. The key of the dictionary is a tuple of two entities (a unipartite edge). The value of the dictionary is the relatedness value.
You can replicate the results of the paper (and see how to use network_map.py and how it works) by using the experiment.py script.
experiment.py requires as input two files: the bipartite network and the unipartite network used for evaluation. In the package, these are the *_bipartite and *_evalnet files respectively, for the four datasets used in the paper (aid_*, congress_*, ipums_* and onet_*). experiment.py will provide as output a ROC plot and the AUC value (in parenthesis, the difference of AUC w.r.t the ycn-rw’s AUC). It will also write several files:
– “x_y_edge” file, containing the unipartite edges. Here, “x” is the dataset name (aid, congress, ipums, onet) and “y” is the projection method name (ycn, zho, jac, euc, pea, cos).
– “x_y_roc” file, containing the data to plot the ROC curve. Again, “x” is the dataset name (aid, congress, ipums, onet) and “y” is the projection method name (ycn, zho, jac, euc, pea, cos).
– “x_auc” file, containing the AUC values. Here, “x” is the dataset name (aid, congress, ipums, onet).
To run experiment.py , make sure that the input files, network_map.py and pyroc.py are in the same folder, then type in your shell: python experiment.py dataset_name significance_threshold, where “dataset_name” is the filename part before the “_bipartite” and “_evalnet” extensions of your input files; and “significance_threshold” is the threshold for your unipartite edges to be considered significant.
For example, to replicate the paper experiments, these are the commands used:
python experiment.py aid 12
python experiment.py congress 7
python experiment.py ipums 7
python experiment.py onet 140
experiment_movielens.py runs with the same set up, without the significance_threshold parameter. It does not generate aucs and rocs, but two files describing the rs and hit rates. See Zhou et al.’s “Bipartite network projection and personal recommendation” for more details about how to interpret the results.
For any inquiry, contact me at: michele.coscia@gmail.com
Happy bipartite network projection!
